pLVX-IRES-tdTomato
Catalog No. PVT11066
Packing 2ug
pLVX-IRES-tdTomato Information
Promoter: CMV
Replicon: pUC ori
Terminator: SV40 poly (A) signal
Plasmid Classification: Virus Series, Lentivirus Cloning Vectors
Plasmid size: 8892 BP
Prokaryotic resistance: Amp
Cloning strain: Stbl3
Culture conditions: 37 C, aerobic LB
Expression host: mammalian cells
Induction mode: no induction, transient expression
5'Sequencing Primers: CMV-F: CGCAAATGGGGGGGTAGGGGGGTG
3'Sequencing primers: WPRE-R: CATAGCGTAAAAGGAGCAACA
pLVX-IRES-tdTomato Description
pLVX-IRES-tdTomato is an HIV-1-based, lentiviral expression vector that allows the simultaneous expression of your protein of interest and tdTomato in virtually any mammalian cell type, including primary cells. tdTomato is a member of the family of fruit fluorescent proteins derived from the Discosoma sp. red fluorescent protein, DsRed . The vector expresses the two proteins from a bicistronic mRNA transcript, allowing tdTomato to be used as an indicator of transduction efficiency and a marker for selection by flow cytometry.Expression of the bicistronic transcript is driven by the constitutively active human cytomegalovirus immediate early promoter (PCMV IE) located just upstream of the MCS. An encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES), positioned between the MCS and tdTomato, facilitates cap-independent translation of tdTomato from an internal start site at the IRES/tdTomato junction . pLVX-IRES-tdTomato contains all of the viral processing elements necessary for the production of replication-incompetent lentivirus, as well as elements to improve viral titer, transgene expression, and overall vector function. The woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) promotes RNA processing events and enhances nuclear export of viral RNA , leading to increased viral titers from packaging cells. In addition, the vector includes a Rev-response element (RRE), which further increases viral titers by enhancing the transport of unspliced viral RNA out of the nucleus . Finally, pLVX-IRES-tdTomato also contains a central polypurine tract/central termination sequence element (cPPT/CTS). During target cell infection, this element creates a central DNA flap that increases nuclear import of the viral genome, resulting in improved vector integration and more efficient transduction. The vector also contains a pUC origin of replication and an E. coli ampicillin resistance gene (Ampr ) for propagation and selection in bacteria.
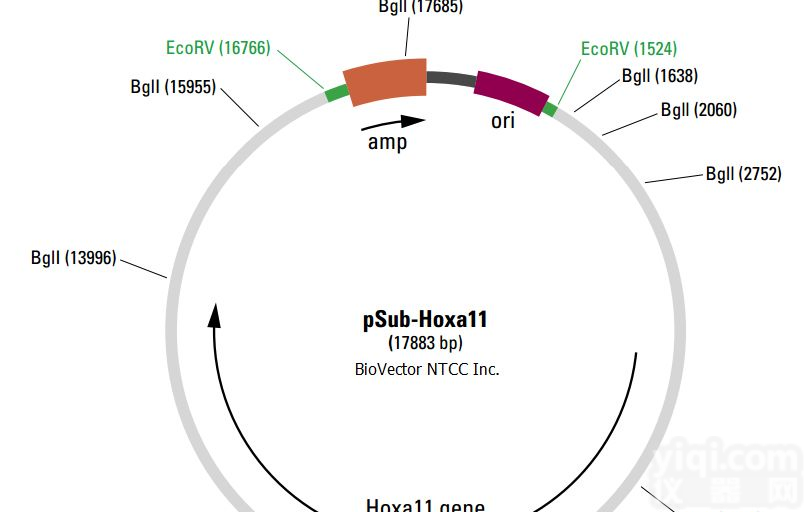
pLVX-IRES-tdTomato Sites

pLVX-IRES-tdTomato Sequence
LOCUS Exported 8892 bp ds-DNA circular SYN 09-SEP-2016
DEFINITION synthetic circular DNA
ACCESSION .
VERSION .
KEYWORDS Untitled 4
SOURCE synthetic DNA construct
ORGANISM synthetic DNA construct
REFERENCE 1 (bases 1 to 8892)
AUTHORS .
TITLE Direct Submission
JOURNAL Exported Friday, September 9, 2016 from SnapGene Viewer 3.1.4
FEATURES Location/Qualifiers
source 1..8892
/organism="synthetic DNA construct"
/mol_type="other DNA"
LTR 1..634
/note="3' LTR"
/note="3' long terminal repeat (LTR) from HIV-1"
misc_feature 681..806
/note="HIV-1 Psi"
/note="packaging signal of human immunodeficiency virus
type 1"
misc_feature 1303..1536
/note="RRE"
/note="The Rev response element (RRE) of HIV-1 allows for
Rev-dependent mRNA export from the nucleus to the
cytoplasm."
misc_feature 2028..2143
/note="cPPT/CTS"
/note="central polypurine tract and central termination
sequence of HIV-1 (lacking the first T)"
enhancer 2201..2504
/note="CMV enhancer"
/note="human cytomegalovirus immediate early enhancer"
promoter 2505..2708
/note="CMV promoter"
/note="human cytomegalovirus (CMV) immediate early
promoter"
misc_feature 2843..3415
/note="IRES"
/note="internal ribosome entry site (IRES) of the
encephalomyocarditis virus (EMCV)"
misc_feature 2866..3417
/note="IRES"
/note="internal ribosome entry site (IRES) of the
encephalomyocarditis virus (EMCV)"
CDS 3417..4847
/codon_start=1
/product="tandem dimeric (pseudo-monomeric) derivative of
DsRed (Shaner et al., 2004)"
/note="tdTomato"
/note="mammalian codon-optimized"
/translation="MVSKGEEVIKEFMRFKVRMEGSMNGHEFEIEGEGEGRPYEGTQTA
KLKVTKGGPLPFAWDILSPQFMYGSKAYVKHPADIPDYKKLSFPEGFKWERVMNFEDGG
LVTVTQDSSLQDGTLIYKVKMRGTNFPPDGPVMQKKTMGWEASTERLYPRDGVLKGEIH
QALKLKDGGHYLVEFKTIYMAKKPVQLPGYYYVDTKLDITSHNEDYTIVEQYERSEGRH
HLFLGHGTGSTGSGSSGTASSEDNNMAVIKEFMRFKVRMEGSMNGHEFEIEGEGEGRPY
EGTQTAKLKVTKGGPLPFAWDILSPQFMYGSKAYVKHPADIPDYKKLSFPEGFKWERVM
NFEDGGLVTVTQDSSLQDGTLIYKVKMRGTNFPPDGPVMQKKTMGWEASTERLYPRDGV
LKGEIHQALKLKDGGHYLVEFKTIYMAKKPVQLPGYYYVDTKLDITSHNEDYTIVEQYE
RSEGRHHLFLYGMDELYK"
misc_feature 4861..5449
/note="WPRE"
/note="woodchuck hepatitis virus posttranscriptional
regulatory element"
CDS complement(5332..5343)
/codon_start=1
/product="Factor Xa recognition and cleavage site"
/note="Factor Xa site"
/translation="IEGR"
LTR 5656..6289
/note="3' LTR"
/note="3' long terminal repeat (LTR) from HIV-1"
primer_bind complement(6417..6433)
/note="M13 rev"
/note="common sequencing primer, one of multiple similar
variants"
protein_bind 6441..6457
/bound_moiety="lac repressor encoded by lacI"
/note="lac operator"
/note="The lac repressor binds to the lac operator to
inhibit transcription in E. coli. This inhibition can be
relieved by adding lactose or
isopropyl-beta-D-thiogalactopyranoside (IPTG)."
promoter complement(6465..6495)
/note="lac promoter"
/note="promoter for the E. coli lac operon"
protein_bind 6510..6531
/bound_moiety="E. coli catabolite activator protein"
/note="CAP binding site"
/note="CAP binding activates transcription in the presence
of cAMP."
rep_origin complement(6819..7407)
/direction=LEFT
/note="ori"
/note="high-copy-number ColE1/pMB1/pBR322/pUC origin of
replication"
温馨提示:不可用于临床ZL。